SNP arrays are enabling tools for high-resolution studies of the genetic basis of complex traits, and for incorporating genetic markers to expedite genetic gain in selective breeding programmes. The development of a SNP array for the European sea bass (Dicentrarchus labrax) and the gilthead sea bream (Sparus aurata) would be of significant importance for Mediterranean aquaculture, as it would allow, among other relevant applications, to implement Genomic Selection (GS).
GS is a method by which genome-wide genetic marker data are used to predict the breeding values of individuals with higher accuracy than pedigree-based methods. Consequently, high-density marker panels have a major potential for accelerating genetic gain of breeding programmes through GS, particularly for traits practically impossible to measure on selection candidates, such as disease resistance.
As the commercial-scale application of GS requires relatively low-cost genome-wide genotyping platforms, adequate platforms need to be developed to fully exploit its benefits. A cost-effective SNP array can be developed by combining SNPs derived from two species onto a single platform. In this way, a significant reduction in genotyping costs can be achieved when higher volumes of units are ordered, which at the same time would facilitate the uptake of the technology by the industry and research community.
As part of the MedAID project we developed MedFish, the first publically available combined-species 60K SNP array for the European sea bass and the gilthead sea bream. The entire process of sample collection, sequencing and bioinformatics was performed together with the PerformFISH project (H2020 agreement No 727610) in order to generate one single platform for widespread use. This partnership between MedAID and PerformFISH ensured that the price of the array was lower due to the higher volume of samples genotyped across the two projects. The collaboration also allowed a broader sampling of more fish populations across Europe, which is likely to widen the applicability of the array for use in future studies and breeding programmes.
The first step we followed to develop the MedFish SNP array was to discover genetic variants in representative farmed and wild European sea bass and gilthead sea bream populations. Farmed populations are considered to be those that were sampled from commercial hatcheries or established farms. To this end, over 24 populations of European sea bass and 27 populations of gilthead sea bream were sampled across 11 Mediterranean countries (Figure 1).
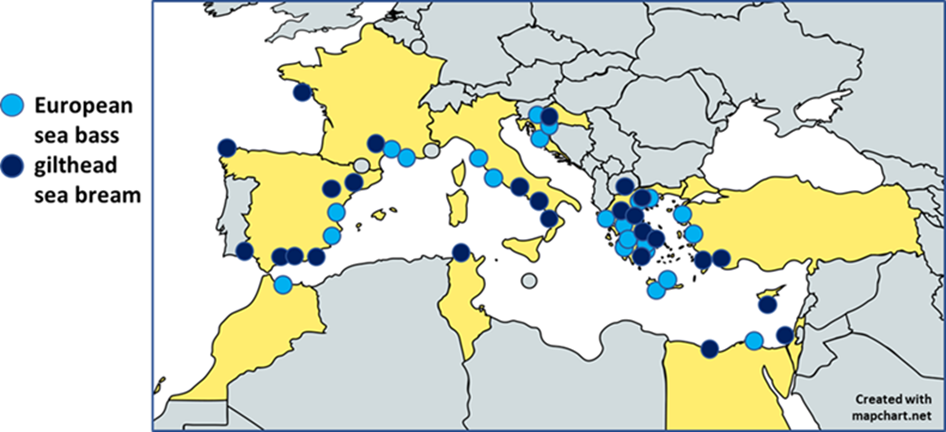
A pooled whole-genome resequencing (Pool-Seq) approach was followed to maximize the number of variants detected in a cost-effective manner. In total, the whole genome of over 500 individuals of each fish species were sequenced on either a HiSeq4000 or a HiSeqX platform. The output sequence data (>3000 Gb) was then processed to discover and characterise a substantial SNP database.
A SNP discovery pipeline (Figure 2) was developed to evaluate the raw sequencing data and identify high confidence SNP variants by aligning short paired-end reads to the reference genomes of the two species. In an initial evaluation of the data, ~17 and ~34 million variants were discovered for the European sea bass and gilthead sea bream, respectively. After filtering for high quality variants, a list of reliable candidate variants was obtained for each species.
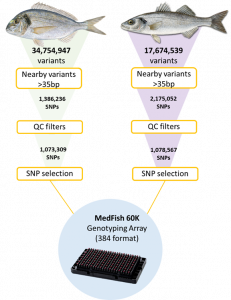
The second step in the development of the technology was the selection of variants for inclusion in the MedFish SNP array. The platform is designed to genotype ~60K SNP markers. Therefore, ~30K markers were selected for each species from the substantial SNP database that was generated from the initial screening of the fish populations. For the process of SNP selection, markers were divided into selection tiers based on their hierarchy of importance. SNP markers that (i) showed an association with production traits in previous studies or (ii) were predicted to have a high effect on genes (e.g. lead to truncated proteins) were considered high priority markers, given their potential relevance for the industry. In addition, a subset of markers from the MedFish SNP array are shared with previous platforms, as a way of ensuring backward compatibility. MedAID and PerformFISH discovered all markers present in the MedFish platform, which cover all European sea bass and gilthead sea bream chromosomes at a density that depends on the estimated local nucleotide diversity, with high minor allele frequency being the main inclusion criteria.
The MedFish SNP array will be applied within the MedAID project to (i) perform genomic analysis of production traits in the European sea bass and the gilthead seabream, (ii) estimate the relatedness of populations and loss of genetic variation in the European seabass and the gilthead seabream, and (iii) study genotype x environment interaction for production efficiency traits in the European sea bass.
This newly generated SNP array will be commercially available to the industry and publicly available to the research community through Thermo Fisher Scientific. We anticipate that this platform will become an essential tool to improve the competitiveness and sustainability of the European sea bass and the gilthead sea bream aquaculture industry throughout the Mediterranean.
Access to the full deliverable