The term “Epigenetics” was first introduced by Conrad H. Waddington (1905–1975) in 1942 [1] to describe events that were found not to be congruent with the genetic principles. Today, epigenetic changes concern changes in the structure of DNA resulting from DNA modifications. Contrary to DNA mutations, epigenetic changes are reversible and occur fast.
Apart from the programmed epigenetic regulation that ensures cell differentiation, epigenetic regulation can be influenced by the environment, emphasizing the importance to investigate in depth epigenetic mechanisms (Fig.1). Especially in terms of climate change, which results in changes of temperature, acidity, and salinity in the marine milieu, epigenetics may shed light on the mechanism of phenotypic response triggered by environmental signals.
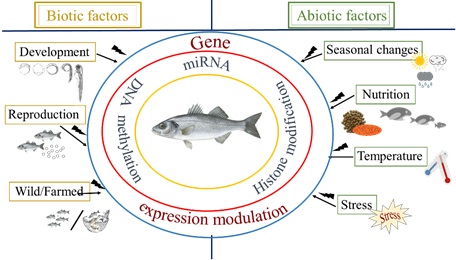
One of the most important environmental parameters influencing gene expression in poikilothermic animals -such as fish- via epigenetic regulators, are temperature fluctuations during early life. Temperature is known to control every biological function in such organisms, especially during early larval development, and can have both direct effects, such as on the growth rate, the development of skeletal deformities and the swimming speed of larvae, but also indirect effects, such as on sex differentiation and the final sex ratio of the populations in fish with temperature-dependent sex determination system (TSD).
Epigenetic regulation may act at the level of transcription that is mediated by histone modification and DNA methylation (Fig. 2). The modification of histones takes place post-translationally (PTM) and may alter gene expression. Histone modification may play an important regulative role in diverse biological processes, such as transcriptional activation, but also inactivation, chromosome packaging and DNA damage/repair. The molecular machinery responsible for histone modifications is largely regulated during embryonic development and may be influenced by environmental conditions [2]. Unlike histone modification, DNA methylation is one of the most studied epigenetic markers. In this case, a methyl group is added to a cytosine residue in DNA, which occurs mostly at CpG islands. It has been reported that DNA methylation in the promotor region of a gene shows repressive functions. However, DNA methylation may also enhance gene expression, depending on its genomic location. In mammals and plants DNA methylation is sensitive to external factors and extensive DNA reprogramming exists during early development [3]. There is a steadily growing interest in studying DNA methylation in model fish species such as zebrafish (Danio rerio) and medaka (Oryzias latipes), but also in non-model species (reviewed in [4]).
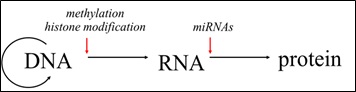
Non-coding RNAs, e.g. miRNAs on the other hand, act at the translational level by mostly repressing the protein production (Fig.2). Non-coding RNAs (ncRNAs) have been shown to be involved in diverse biogenesis pathways and regulatory mechanisms. Among several other types of non-coding regulatory RNAs two main types can be distinguished: long non-coding RNAs (lnc) and small non-coding RNAs. Small non-coding RNAs comprises i) short interfering RNAs (siRNAs), ii) piwi acting RNAs (piRNAs) and iii) microRNAs (miRNAs). The latter are known to be a highly conserved class of small regulatory RNAs that essentially contribute to regulatory growth and adaptive plasticity networks. The majority of studies on miRNAs reveal repressive role on the post-transcriptional expression of target mRNAs as their primary function [5]. MiRNAs involved in fish development have been first described in zebrafish [5]. Likewise, miRNAs in teleost gonads have been reported in model fish species, as well as in some of the commercially important fish species [6]. With the commencement of next generation sequencing (NGS) and advanced computational and laboratory technologies, the discovery of new miRNAs from other, non-model species, such as the Atlantic halibut (Hippoglossus hippoglossus) [7,8] and the European sea bass (Dicentrarchus labrax) has been enhanced greatly [9]. The identification of miRNAs in fish showed conservation of their sequence throughout vertebrate species. In addition, the conservation of miRNA sequence has expanded in the recognition of specific target mRNAs, as well as in their involvement in particular biological mechanisms [10,11].
Epigenetic regulation of gene expression by temperature has been correlated to both DNA methylation [12–14] and miRNAs [15] in different fish species, including the European sea bass, one of the most important Mediterranean aquaculture fish species and the target species in MedAID. Previous studies in the European sea bass have shown the existence of TSD [16], resulting in an excess of males in aquaculture populations when high temperatures are used [17–20]. Besides the significance of sex control in aquaculture influenced by environmental factors, embryonic and larval stages represent one of the most critical periods to ensure high performance and superior quality in the subsequent developmental phases of the life cycle. Key developmental events occur early in development and are influenced by external parameters such as stress, temperature, salinity and photoperiod. Any failure may cause malformations, developmental delays, poor growth and massive mortalities.
While the significance in studying epigenetic mechanisms has been widely acknowledged, so far studies have been mainly carried out in agricultural and livestock science, with emphasis on DNA methylation and miRNAs. Epigenetic studies in economically important aquaculture species, however, are still in their infancy although many aquaculture operations are exposed to a broad range of environmental -and at times unnatural- changes. Especially in marine aquaculture, it has been acknowledged that one of the areas of emphasis is the effects of environmental parameters on early rearing, growth, nutrition and health. This has also been recognized by several research projects, including MedAID, funded by the EU in terms of generating epigenetic markers for aquaculture (e.g. ERC consolidator grant EPIFISH: Innovative epigenetic markers for fish domestication, JMO Fernandes).
![]() Complete article with references
Complete article with references

Authors: E. Sarropoulou, E. Kaitetzidou, M. Papadaki and C.C. Mylonas,
Hellenic Center for Marine Research, Crete, Greece.

